A Multi-cell, Multi-scale Model of Vertebrate Segmentation and Somite Formation
How can a densely packed PSM tissue mechanically split up into a string of separate somites?
Introduction
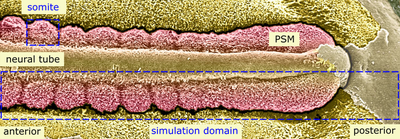
Somitogenesis is the defining developmental process of vertebrate species and essential for the segmentation of the main body axis and, among other repetitive structures, the formation of vertebrae. Early in vertebrate embryo development, individual segments known as somites sequentially bud off the presomitic mesoderm (PSM), pairwise on either side of the notochord. This periodic process is controled by the phases of a molecular oscillator, termed the segmentation clock, in the PSM cells. Clock phases affect gene expression, including adhesion proteins of the Eph and ephrin families, and differentiation of cellular subpopulations within each presumptive somite. An important question, addressed by the presented model, is how the densely packed PSM tissue mechanically splits up into a string of separate somites on each side of the body axis.
The Clock-Wavefront Model
To read out the phase of the segmentation clock at a specific moment, Cooke and Zeeman in 1976 had proposed the clock-wavefront model, i.e. a threshold value of a morphogen gradient to impose the moment of clock read-out. The morphogene gradient would move posteriorly with PSM outgrowth such that the threshold value gives a wavefront.
The presented model includes a field of Fibroblast growth factor 8 (FGF8) as the morphogen. Produced and secreted by the PSM cells, extracellular FGF8 diffuses freely and is degraded. Cells sense their local FGF8 concentration and compare it to a specific threshold value. When both match then the differentiation process initiates, contingent on the state (molecular concentrations representing the phase) of the intracellular segmentation clock.
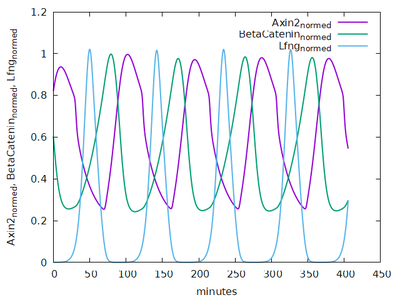
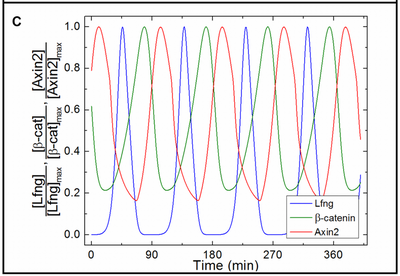
The segmentation clock is modeled as a system of ODEs for the concentrations of signaling molecules in each PSM cell. Delta-Notch signaling couples any PSM cell to the states of all its neighbors. Though this clock model comprises 20 ODEs and 52 parameters, only three variables are crucial for read-out: lunatic fringe protein (Lfng) concentration, Axin2 concentration, and Beta-Catenin concentration. Each variable dominates one of three clock phases. These three concentration values at the wavefront determine the differentiation process. Consequently, somites in this model become composed of three distinct cell layers: top, core, and bottom.
Details and parameter values of the presented model had been adjusted in the cited publication for a chick embryo.
Model Description
The cited publication thoroughly documents the model which was developed with the software CompuCell3D and the model files were shared with the publication, see Text S2.
The model replication in Morpheus, as presented here, exactly follows the paper’s detailed model description.
This is possible for CPM-based models as both simulators provide solvers for the CPM.
The technical settings of node neighborhoods on the lattice for surface estimation and CPM node updates are identical between CompuCell3D and Morpheus when the latter is run in surface scaling mode classic, i.e. surface length with the fixed 1st order neighborhood kernel but contact energies with the chosen neighborhood kernel, here 2nd order.
In the following, key aspects of the original model (and likewise of its replication) are summarized.
The model considers a 2D cross section of the embryo and just one half of the left-right symmetric process, hence a single string of somites. The 2D square lattice is spanned by the anterior-posterior body axis (top-down in following images) and the medio-lateral axis (left-right).
Length and time units are set as follows: a lattice node has a length of 1.43 µm and a model time step is 1 MCS and corresponds to 0.015 min. As one somite in chick is formed every 90 min, this occurs every 6000 time steps in the model.
Initial Conditions
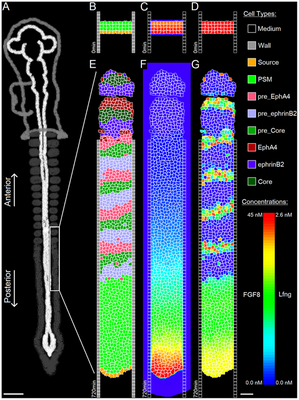
The model starts with two columns of 150 immobile wall cells each, maintaining the PSM’s shape. The domain’s top corresponds to anterior and bottom to posterior. At the top, we initialize three layers of PSM cells, with a layer of Source cells underneath (these cell type names as literally used in the computational model are distinguished by code highlighting).
PSM Growth
In this model, only the (artificial layer of) Source cells divide such that normal (linear) PSM growth results. Each Source cell grows (target volume and target surface are gradually increased and CPM updates let the cell shape follow these targets) at a rate set by the parameter gR=0.120 in model section Global. Once a Source cell has exceeded twice its initial cell volume, it divides. Daughter cells that stay at the tissue edge where they keep contact with the medium stay Source cells while the other daugther cell when it gets surrounded by other cells will become a PSM cell. This division model, combined with a ConnectivityConstraint for unfragmented cell shapes, ensures a consistent layer of Source cells and supplies new PSM cells at a constant rate. Thus, the PSM grows towards the posterior end while individual cells move relatively little on random walks.
FGF8 Field
The FGF8 concentration is modeled via a reaction-diffusion PDE:
with uniform and isotropic diffusion and degradation rate k. The source term PSM cell and Source cell produces FGF8 and secretes it. This is modeled by assigning a fixed amount of mRNA (that encodes for FGF8) to Source cells and by computing for each PSM cell a concentration of mRNA. Abstracting many biological details, the FGF8 secreted by a cell is set proportional to its current mRNA amount. Intracellular mRNA decays exponentially (except in Source cells) at a considerably smaller rate than the extracellular FGF8 decays.
Combined with PSM growth, this leads to the formation of a spatial FGF8 gradient that moves posterior as the PSM grows.
Source cells. Right: Submodel for PSM growth and FGF8 dynamics. To obtain an FGF8 gradient that spans the entire anterior-posterior axis, FGF8 secretion has to remain active in determined cells.
Clock-dependend Determination and Differentiation
As described in the Introduction, the differentiation is triggered by reaching a certain FGF8 threshold and specified by the state of the segmentation clock. The implementation for the clock as a coupled system of ODEs is straight forward, both in CompuCell3D and Morpheus. The sensing of extracellular FGF8 concentration occurs at a single lattice node at the cell’s center of mass, both in CompuCell3D and Morpheus.
The differentiation process is modeled as follows: When a cell hits the FGF8 determination threshold, it transitions to a “determined cell,” an intermediary state between PSM cells and the fully differentiated cells. Once determined, a cell’s final type among the three somitic cell layers is set with a delay. This intermediate step is necessary for cell-sorting, hence, already differentiated cells and determined cells differ in their contact energies.
Here, time courses of the three clock outputs from an isolated cell with fixed FGF8 and Wnt3 input values are compared to the published results.
Cell-Cell Interactions and Somite Formation
The model comprises 8 biologically motivated cell types: Source, PSM, preEphA4, preEphrinB2, preCore, EphA4, EphrinB2, Core.
Contact energies are defined for each pair of cell types.
Results
As in the cited publication, the modularity of the model allows to analyze and compare results of its separated submodels (see figures and videos above), now followed by the complete model dynamics.
Here, all submodels are coupled by feedback loops and individual cells yield different dynamics in response to different inputs according to their local neighborhood and position (cell-cell adhesion, Delta-Notch signals from cell-cell contacts, diffusible FGF signal, Wnt signal gradient). These signals synchronize clock phases in nearby cells and generate a spatial profile of the clock period, that becomes visible as a phase wave.
Complete Model
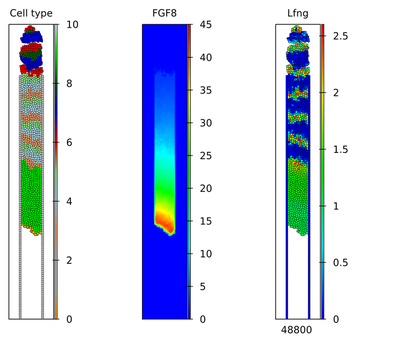
As the submodels successfully reproduced the published results, also the complete model in Morpheus does so:
The published simulation results were presented in separate Videos S1 and S2, as shown below:
These simulation results can be compared for individual time points as shown below.
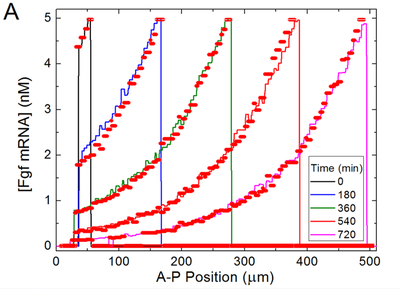
Profile of Oscillation Period
Hester et al. observed an emergent increase of the segmentation clock period from the posterior end towards anterior. We observed the same behavior both qualitatively and quantitatively in the separate model file submodel_clock.xml that omits downstream differentiation processes as in the original model. These Morpheus results (panels C,D) match the original (panels A,B) as compared in the following figure:
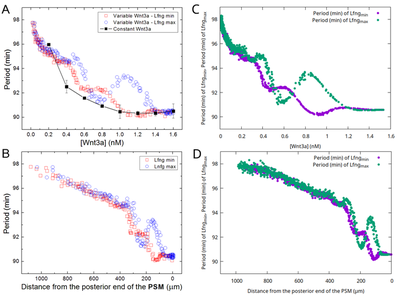
We reproduced the figure in Morpheus (even measuring the temporal period and constructing the entire plot on the fly in the running model) by measuring in each PSM cell the period length both between subsequent maxima and minima of the Lfng time course, respectively. The slight data shift towards lower Wnt3a in panel (C) compared to (A) resulted from plotting against the lower Wnt3a values at the end of each clock period (in C) instead of the mean Wnt concentration (in A).
Note that daughters of source cells here inherited the clock phase of the mother cell and thereby started in sync. The incorporated Delta-Notch pathway may also dynamically synchronize initially random clock phases, which could be an interesting follow-up question. The supplemental model code as documented in the appendix of S. Hester’s dissertation included the possibility to randomize the clock phase upon cell division (see page 129 and page 149). In the Morpheus model, such phase variability could also be introduced by running the Systems with Clock ODEs individually faster or slower (using the Scaling attribute) in daughter cells for some time interval.
In addition to the matching videos above, this reproduction of the period profiles is very sensitive to details in all coupled submodels and confirms the published results in an independent simulator.
Reference
This model reproduces a published result, originally obtained with a different simulator:
S. D. Hester, J. M. Belmonte, J. S. Gens, S. G. Clendenon, J. A. Glazier: A Multi-cell, Multi-scale Model of Vertebrate Segmentation and Somite Formation. PLoS Comput. Biol. 7 (10): e1002155, 2011.
Model
model_main.xml
XML Preview
<?xml version='1.0' encoding='UTF-8'?>
<MorpheusModel version="4">
<Description>
<Title>Somitogenesis</Title>
<Details>Full title: A Multi-cell, Multi-scale Model of Vertebrate Segmentation and Somite Formation
Date: 18.08.2023
Authors: Susann D. Hester, Julio M. Belmonte, J. Scott Gens, Sherry G. Clendenon, James A. Glazier
Curators: Justin Buerger, Lutz Brusch
Software: Morpheus (open source). Download from: https://morpheus.gitlab.io
Model ID: https://identifiers.org/morpheus/M6696
Units: [time] = 0.015 min
[space] = 1.43 µm
This model reproduces the model and results obtained with CompuCell3D in the original publication.
Reference:
Susann D. Hester, Julio M. Belmonte, J. Scott Gens, Sherry G. Clendenon, James A. Glazier: A Multi-cell, Multi-scale Model of Vertebrate Segmentation and Somite Formation. PLoS Comput. Biol. 7(10): e1002155, 2011.
https://doi.org/10.1371/journal.pcbi.1002155
</Details>
</Description>
<Space>
<Lattice class="square">
<Neighborhood>
<Order>4</Order>
</Neighborhood>
<Size symbol="size" value="2*margin + 2*wall + 10*cD, 145*cD, 0.0"/>
<BoundaryConditions>
<Condition type="noflux" boundary="x"/>
<Condition type="noflux" boundary="-x"/>
<Condition type="noflux" boundary="y"/>
<Condition type="noflux" boundary="-y"/>
</BoundaryConditions>
</Lattice>
<SpaceSymbol symbol="space"/>
</Space>
<Time>
<StartTime value="0"/>
<StopTime value="120000"/>
<TimeSymbol symbol="time"/>
</Time>
<Analysis>
<ModelGraph include-tags="#untagged" format="dot" reduced="false"/>
<Gnuplotter time-step="200">
<Terminal name="png" size="1600, 1600, 0"/>
<Plot title="Cell type">
<Cells value="cell.type">
<ColorMap adaptive-range="false">
<Color color="orange" value="0"/>
<Color color="gray" value="1"/>
<Color color="gray" value="2"/>
<Color color="light-salmon" value="3"/>
<Color color="light-blue" value="4"/>
<Color color="light-green" value="5"/>
<Color color="red" value="6"/>
<Color color="blue" value="7"/>
<Color color="dark-green" value="8"/>
<Color color="green" value="9"/>
<Color color="white" value="10"/>
</ColorMap>
</Cells>
</Plot>
<Plot title="FGF8">
<Field symbol-ref="FGF" max="45" min="0">
<ColorMap adaptive-range="false">
<Color color="red" value="45"/>
<Color color="yellow" value="33.75"/>
<Color color="green" value="22.5"/>
<Color color="cyan" value="11.25"/>
<Color color="blue" value="0"/>
</ColorMap>
</Field>
</Plot>
<Plot title="Lfng">
<Cells max="2.6" min="0" value="F">
<ColorMap adaptive-range="false">
<Color color="red" value="2.6"/>
<Color color="yellow" value="1.95"/>
<Color color="green" value="1.3"/>
<Color color="cyan" value="0.65"/>
<Color color="blue" value="0"/>
</ColorMap>
</Cells>
</Plot>
</Gnuplotter>
</Analysis>
<CellTypes>
<CellType name="Source" class="biological">
<CellDivision daughterID="daughter" division-plane="oriented" orientation="0.0, -1.0, 0.0">
<Condition>cell.volume >= 2*tV</Condition>
<Annotation>Cell Division: as soon as the source cell reaches double its default target volume</Annotation>
<Triggers>
<Rule symbol-ref="targetVolume">
<Expression>tV</Expression>
</Rule>
<Rule symbol-ref="targetSurface">
<Expression>tS</Expression>
</Rule>
</Triggers>
</CellDivision>
<ChangeCellType time-step="1" newCellType="PSM" name="to PSM">
<Condition>MediumNeighbor == 0</Condition>
</ChangeCellType>
<Property symbol="targetVolume" value="tV"/>
<Property symbol="targetSurface" value="tS"/>
<System time-step="1.0" name="Source cell growth" solver="Euler [fixed, O(1)]">
<Rule symbol-ref="targetVolume">
<Expression>if((targetVolume < 3*tV), (targetVolume+gR), targetVolume)</Expression>
</Rule>
<Rule symbol-ref="targetSurface">
<Expression>if((targetVolume < 3*tV), 5*sqrt(targetVolume), targetSurface)</Expression>
</Rule>
</System>
<VolumeConstraint target="targetVolume" strength="LamV"/>
<SurfaceConstraint target="targetSurface" strength="LamS" mode="surface"/>
<Property symbol="MediumNeighbor" value="0.0"/>
<NeighborhoodReporter>
<Input scaling="cell" value="cell.type == celltype.Medium.id"/>
<Output symbol-ref="MediumNeighbor" mapping="sum"/>
</NeighborhoodReporter>
<Mapper time-step="1.0" name="sense FGF8 at cell center">
<Input value="if((space.x-1<=cell.center.x && space.x+1>=cell.center.x && space.y-1<=cell.center.y && space.y+1>=cell.center.y),FGF,0)"/>
<Output symbol-ref="FGF_center" mapping="maximum"/>
</Mapper>
<Property symbol="FGF_center" value="45"/>
<Property symbol="N" name="Notch in the membrane" value="0.1144"/>
<Property symbol="N_a" name="activated Notch" value="0.7745"/>
<Property symbol="N_an" name="Activated Notch in the nucleus" value="0.00243"/>
<Property symbol="N_ap" name="phosphorylated notch" value="0.0013"/>
<Property symbol="MF" name="lunatic fringe mRNA" value="1.2464"/>
<Property symbol="F" name="lunatic fringe protein" value="2.3795"/>
<Property symbol="MDMF" name="delta modification factor mRNA" value="0.0011"/>
<Property symbol="DMF" name="delta modification factor protein" value="0.2090"/>
<Property symbol="DL_c" name="delta protein in cytoplasm" value="0.1255"/>
<Property symbol="DL_m" name="delta protein in the membrane" value="0.0906"/>
<Property symbol="DL_mExt" name="Delta outside the cell" value="DL_m"/>
<Property symbol="Sum_DL_mExt" name="Delta outside the cell" value="DL_m"/>
<Property symbol="PSM_Neighbors" name="Delta outside the cell" value="1"/>
<NeighborhoodReporter time-step="1.0" name="Delta/Notch from neighboring cells">
<Input scaling="cell" value="DL_m"/>
<Output symbol-ref="Sum_DL_mExt" mapping="sum"/>
</NeighborhoodReporter>
<NeighborhoodReporter time-step="1.0" name="Neighboring PSM cells">
<Input scaling="cell" value="(cell.type == celltype.Source.id) + (cell.type == celltype.PSM.id) + (cell.type == celltype.preEphA4.id) + (cell.type == celltype.preCore.id) + (cell.type == celltype.preEphrinB2.id)"/>
<Output symbol-ref="PSM_Neighbors" mapping="sum"/>
</NeighborhoodReporter>
<System time-step="1.0" name="Delta/Notch signaling" solver="Euler [fixed, O(1)]">
<Rule symbol-ref="DL_mExt">
<Expression>if(PSM_Neighbors != 0, Sum_DL_mExt/PSM_Neighbors, 0)</Expression>
</Rule>
</System>
<System time-step="1.0" name="Notch loop" solver="Runge-Kutta [fixed, O(4)]">
<DiffEqn symbol-ref="N">
<Expression>eps*(mu_sN-mu_dN*N/(K_dN+N)-(k_c*N*(K_IF^2/(K_IF^2 + F^2))*((DL_mExt/(K_DL +DL_mExt))/(K_aDL + (DL_mExt/(K_DL +DL_mExt))))))</Expression>
</DiffEqn>
<DiffEqn symbol-ref="N_a">
<Expression>eps*((k_c*N*(K_IF^2/(K_IF^2 + F^2))*((DL_mExt/(K_DL +DL_mExt))/(K_aDL + (DL_mExt/(K_DL +DL_mExt)))))-mu_dNa*N_a/(K_dNa +N_a)-(k_t1*N_a-k_t2*N_an))</Expression>
</DiffEqn>
<DiffEqn symbol-ref="N_an">
<Expression>eps*((k_t1*N_a-k_t2*N_an)-mu_dNan*N_an/(K_dNan + N_an))</Expression>
</DiffEqn>
<DiffEqn symbol-ref="MF">
<Expression>eps*(mu_sF*(N_an-(mu_Nap*N_an*K/(K_pN+K)))^2/(KA^2 +(N_an-(mu_Nap*N_an*K/(K_pN+K)))^2)-mu_mF*MF/(K_dmF+MF))</Expression>
</DiffEqn>
<DiffEqn symbol-ref="F">
<Expression>eps*(k_sF*MF -mu_dF*F/(K_dF+F))</Expression>
</DiffEqn>
<DiffEqn symbol-ref="MDMF">
<Expression>eps*(mu_s0MDMF*(N_an-(mu_Nap*N_an*K/(K_pN+K)))^2/(K_aNDMF^2+(N_an-(mu_Nap*N_an*K/(K_pN+K)))^2) - mu_dmDMF*MDMF/(K_dmDMF +MDMF))</Expression>
</DiffEqn>
<DiffEqn symbol-ref="DMF">
<Expression>eps*(k_sDMF*MDMF - mu_dDMF*DMF/(K_dDMF + DMF))</Expression>
</DiffEqn>
<DiffEqn symbol-ref="DL_c">
<Expression>eps*(k_sDL*MDL*(N_an -(mu_Nap*N_an*K/(K_pN+K)))/(K_Nan +(N_an-(mu_Nap*N_an*K/(K_pN+K)))) -k_tDL*DL_c - mu_dDLc*DL_c/(K_dDLc + DL_c))</Expression>
</DiffEqn>
<DiffEqn symbol-ref="DL_m">
<Expression>eps*(k_tDL*DL_c - mu_dDLm*DL_m/(K_dDLm + DL_m))</Expression>
</DiffEqn>
<Constant symbol="mu_sN" name="maximum rate of notch synthesis" value="0.23*mpm"/>
<Constant symbol="mu_dN" name="maximum rate of notch degradation" value="2.82*mpm"/>
<Constant symbol="K_dN" name="michaelis constant for notch degradation" value="1.4"/>
<Constant symbol="k_c" name="rate for notch cleavage" value="3.45*mpm"/>
<Constant symbol="mu_dNa" name="maximum rate of cytoplasmic NICD degradation" value="0.01*mpm"/>
<Constant symbol="K_dNa" name="michaelis constant for cytoplasmic NICD degradation" value="0.001"/>
<Constant symbol="mu_dNan" name="maximum rate of nuclear NICD degradation" value="0.1*mpm"/>
<Constant symbol="K_dNan" value="0.001"/>
<Constant symbol="K_IF" name="threshold constant for inhibition of notch cleavage by Lfng" value="0.45"/>
<Constant symbol="K_aDL" name="delta signaling threshold constant for notch cleavage" value="0.035"/>
<Constant symbol="k_t1" name="rate constant for NICD entry into nucleus" value="0.1*mpm"/>
<Constant symbol="k_t2" value="0.1*mpm"/>
<Constant symbol="mu_Nap" name="maximum fraction of NICD phosphorylated" value="1.0"/>
<Constant symbol="K_pN" name="Threshold constant for Gsk3Beta-mediated NICD phosphorylation" value="2.5"/>
<Constant symbol="mu_sF" name="maximum rate of Lfng transcription" value="3.24*mpm"/>
<Constant symbol="K_A" name="threshold constant for activation of Lfng gene by NICD" value="0.05"/>
<Constant symbol="mu_mF" name="maximum rate of Lfng mRNA degradation" value="1.92*mpm"/>
<Constant symbol="K_dmF" name="michaelis constant for Lfng mRNA degradation" value="0.768"/>
<Constant symbol="k_sF" name="rate constant for Lfng protein synthesis" value="0.3*mpm"/>
<Constant symbol="mu_dF" name="maximum rate of Lfng protein degradation" value="0.39*mpm"/>
<Constant symbol="K_dF" name="michaelis constant for Lfng protein degradation" value="0.37"/>
<Constant symbol="mu_s0MDMF" name="maximum rate of DMF transcription" value="0.497*mpm"/>
<Constant symbol="K_aNDMF" name="michaelis constant for notch activation of DMF" value="0.05"/>
<Constant symbol="mu_dmDMF" name="maximum rate of DMF mRNA degradation" value="0.314*mpm"/>
<Constant symbol="K_dmDMF" name="michaelis constant for DMD mRNA degradation" value="0.4"/>
<Constant symbol="k_sDMF" name="rate constant for translation of DMF" value="0.1047*mpm"/>
<Constant symbol="mu_dDMF" name="maximum rate of DMF protein degradation" value="0.209*mpm"/>
<Constant symbol="K_dDMF" name="michaelis constant for DMF protein degradation" value="0.5"/>
<Constant symbol="k_sDL" name="rate constant for delta protein synthesis" value="0.75*mpm"/>
<Constant symbol="MDL" name="concentration of delta mRNA" value="0.5"/>
<Constant symbol="K_Nan" name="michaelis constant for NICD stimulation of delta translation" value="0.04"/>
<Constant symbol="k_tDL" name="rate constant for delta protein transport to membrane" value="0.5*mpm"/>
<Constant symbol="mu_dDLc" name="maximum rate of delta proteind egradation in cytoplasm" value="0.5*mpm"/>
<Constant symbol="K_dDLc" name="michaelis constant for delta protein degradation in cytoplasm" value="0.5"/>
<Constant symbol="mu_dDLm" name="maximum rate of delta protein degradation on membrane" value="0.5*mpm"/>
<Constant symbol="K_dDLm" name="michaelis constant for delta protein degradation on membrane" value="0.5"/>
<Constant symbol="K_DL" name="Threshold for delta signaling" value="0.08"/>
<Constant symbol="eps" name="scaling factor for notch loop" value="0.43"/>
<Constant symbol="KA" value="0.05"/>
<Annotation>ODEs for the Notch Loop</Annotation>
</System>
<Property symbol="Ras_a" name="Activated Ras protein" value="1.9888"/>
<Property symbol="ERK_a" name="Activated ERK protein" value="0.01930"/>
<Property symbol="X_a" name="Activated transcription factor X" value="1.9777"/>
<Property symbol="MDusp" name="Dusp mRNA" value="2.1651"/>
<Property symbol="Dusp" name="Dusp protein" value="8.5078"/>
<Constant symbol="mFgf" name="FGF8 mRNA" value="5"/>
<System time-step="1.0" name="FGF loop" solver="Runge-Kutta [fixed, O(4)]">
<DiffEqn symbol-ref="Ras_a">
<Expression>eta*(V_MaRas*((FGF_center^2/(K_aFgf^2 + FGF_center^2))*((Ras_t-Ras_a)/(K_aRas + (Ras_t-Ras_a)))-V_MdRas*(Ras_a/(K_dRas+Ras_a))))</Expression>
</DiffEqn>
<DiffEqn symbol-ref="ERK_a">
<Expression>eta*(V_MaErk*(Ras_a/Ras_t)*(ERK_t-ERK_a)/(K_aErk + (ERK_t-ERK_a))-K_cDusp*Dusp*ERK_a/(K_dErk+ERK_a))</Expression>
</DiffEqn>
<DiffEqn symbol-ref="X_a">
<Expression>eta*(V_MaX*ERK_a/ERK_t*(X_t-X_a)/(K_aX +(X_t-X_a))-V_MdX*X_a/(K_dX + X_a))</Expression>
</DiffEqn>
<DiffEqn symbol-ref="MDusp">
<Expression>eta*(V_MsMDusp*(X_a^2/(K_aMDusp^2 + X_a^2))*(mu_DuspDMF*K_IMDusp/(K_IMDusp +DMF) +mu_DuspX) - V_MdMDusp*MDusp/(K_dMDusp +MDusp))</Expression>
</DiffEqn>
<DiffEqn symbol-ref="Dusp">
<Expression>eta*(K_sDusp*MDusp-V_dDusp*Dusp/(K_dDusp + Dusp))</Expression>
</DiffEqn>
<Constant symbol="Ras_t" name="Total concentration of Ras protein" value="2.0"/>
<Constant symbol="V_MaRas" name="Maximum rate of Ras activation" value="4.968*mpm"/>
<Constant symbol="K_aFgf" name="FGF8 threshold constant for activation of Ras" value="0.5"/>
<Constant symbol="K_aRas" name="Inactie Ras threshold constant for activation of Ras" value="0.103"/>
<Constant symbol="V_MdRas" name="Maximum rate of Ras activation" value="0.41*mpm"/>
<Constant symbol="K_dRas" name="Michaelis constant of Ras activation" value="0.1"/>
<Constant symbol="ERK_t" name="Total concentration of ERK protein kinase" value="2.0"/>
<Constant symbol="V_MaErk" name="Maximum rate of Ras-mediated ERK activation" value="3.3*mpm"/>
<Constant symbol="K_aErk" name="Inactive ERK threshold constant for ERK activation" value="0.05"/>
<Constant symbol="K_cDusp" name="Rate constant for inactivation of ERK" value="1.35*mpm"/>
<Constant symbol="K_dErk" name="Michaelis constant for inactivation of ERK" value="0.05"/>
<Constant symbol="X_t" name="Total concentration of factor X" value="2.0"/>
<Constant symbol="V_MaX" name="Maximum rate of ERK-activated X activation" value="1.6*mpm"/>
<Constant symbol="K_aX" name="Threshold constant for ERK-mediated X activation" value="0.05"/>
<Constant symbol="V_MdX" name="Maximum rate of X inactivation" value="0.5*mpm"/>
<Constant symbol="K_dX" name="Michaelis constant for X inactivation" value="0.05"/>
<Constant symbol="V_MsMDusp" name="maximum rate of Dusp6 transcription" value="0.9*mpm"/>
<Constant symbol="K_aMDusp" name="Threshold constant for X-activated Dusp6-transcription" value="0.5"/>
<Constant symbol="V_MdMDusp" name="maximum rate of Dusp6 mRNA degradation" value="0.5*mpm"/>
<Constant symbol="K_dMDusp" name="Michaelis constant for Dusp6 mRNA degradation" value="0.5"/>
<Constant symbol="K_sDusp" name="rate constant for Dusp6 translation" value="0.5*mpm"/>
<Constant symbol="V_dDusp" name="Maximum rate of Dusp6protein degradation" value="2.0*mpm"/>
<Constant symbol="K_dDusp" name="Michaelis constant for Dusp6 protein degradation" value="0.5"/>
<Constant symbol="mu_DuspX" name="Fraction of Dusp6 transcription solely regulated by X" value="0.2"/>
<Constant symbol="mu_DuspDMF" name="Fraction of Dusp6 transcription under regulation by DMF" value="0.8"/>
<Constant symbol="K_IMDusp" name="Threshold constant for DMF-mediated inhibition of Dusp6" value="0.3"/>
<Constant symbol="eta" name="Scaling factor for FGF8 loop" value="0.328"/>
<Annotation>ODEs for the FGF Loop</Annotation>
</System>
<Property symbol="K" name="Free Gsk3 protein" value="1.4428"/>
<Property symbol="B" name="Beta-Katenin" value="0.4705"/>
<Property symbol="B_p" name="Phosphorylated beta catenin" value="0.0090"/>
<Property symbol="B_n" value="0.2146"/>
<Property symbol="MAx" name="Axin2 mRNA" value="0.1523"/>
<Property symbol="A" name="Axin2 protein" value="0.0461"/>
<System time-step="1.0" name="Wnt loop" solver="Runge-Kutta [fixed, O(4)]">
<DiffEqn symbol-ref="K">
<Expression>teta*(d_1*(K_t-K)-a_1*A*K)</Expression>
</DiffEqn>
<DiffEqn symbol-ref="B">
<Expression>teta*(mu_sB-(V_MK*K_ID/(K_ID+Wnt)*B/(K_1+B))*(K_t-K)/K_t+(V_MP*B_p/(K_2+B_p))+(k_t4*B_n-k_t3*B)-k_d1*B)</Expression>
</DiffEqn>
<DiffEqn symbol-ref="B_p">
<Expression>teta*((V_MK*K_ID/(K_ID+Wnt)*B/(K_1+B))*(K_t-K)/K_t-(V_MP*B_p/(K_2+B_p))-k_d2*B_p)</Expression>
</DiffEqn>
<DiffEqn symbol-ref="B_n">
<Expression>-teta*(k_t4*B_n-k_t3*B)</Expression>
</DiffEqn>
<DiffEqn symbol-ref="MAx">
<Expression>teta*(mu_0+mu_MB*B_n^2/(K_aB^2+B_n^2)+mu_MXa*X_a^2/(K_aXa^2+X_a^2)-mu_md*MAx/(K_md+MAx))</Expression>
</DiffEqn>
<DiffEqn symbol-ref="A">
<Expression>teta*(k_sAx*MAx-mu_dAx*A/(K_dAx+A)+(d_1*(K_t-K)-a_1*A*K))</Expression>
</DiffEqn>
<Constant symbol="a_1" name="Rate constant for Gsk3-Axin-Binding" value="1.8*mpm"/>
<Constant symbol="d_1" name="Rate constant for Gsk3-Axin dissociation" value="0.1*mpm"/>
<Constant symbol="mu_sB" name="Maximum rate of beta-catenin synthesis" value="0.087*mpm"/>
<Constant symbol="k_t3" name="Rate constant for beta-catenin entry into nucleus" value="0.7*mpm"/>
<Constant symbol="k_t4" name="Rate constant for beta-catenin exit from nucleus" value="1.5*mpm"/>
<Constant symbol="V_MK" name="Maximum rate of Gsk3Beta-medaited beta-catein phosphorylation" value="4.5*mpm"/>
<Constant symbol="K_t" name="Total Gsk3Beta concentration" value="3.0"/>
<Constant symbol="K_ID" name="Threshold constant for Dsh inhibition of beta-catenin phosphorylation" value="0.5"/>
<Constant symbol="K_1" name="Michaelis constant for beta-catenin phosphorylation" value="0.28"/>
<Constant symbol="V_MP" name="Maximum rate of beta-catenin dephosphorylation" value="1.0*mpm"/>
<Constant symbol="K_2" name="Michaelis constant for b-catenin dephosphorylation" value="0.03 "/>
<Constant symbol="k_d1" name="Rate constant for unphosphorylated b-catenin degradation" value="0.0*mpm"/>
<Constant symbol="k_d2" name="rate constant for phosphorylated beta-catenin degradation" value="7.062*mpm"/>
<Constant symbol="mu_0" name="Basal rate of Axin2 transcription" value="0.06*mpm"/>
<Constant symbol="mu_MB" name="Maximum rate of beta-catenin activated Axin2 transcription" value="1.64*mpm"/>
<Constant symbol="K_aB" name="Threshold constant for beta-catenon activation of Axin2" value="0.7"/>
<Constant symbol="mu_md" name="Maximum rate of Axin2 mRNA degradation" value="0.8*mpm"/>
<Constant symbol="K_md" name="Michaelis constant for Axin2 mRNA degradation" value="0.48"/>
<Constant symbol="mu_MXa" name="Maximum rate of Xa activated Axin2 transcription" value="0.5*mpm"/>
<Constant symbol="K_aXa" name="Threshold constant for Xa activation of Axin2" value="0.05"/>
<Constant symbol="k_sAx" name="rate constant for axin2 translation" value="0.02*mpm"/>
<Constant symbol="mu_dAx" name="Maximum rate of Axin2 protein degradation" value="0.6*mpm"/>
<Constant symbol="K_dAx" name="Michaelis constant for Axin2 protein degradation" value="0.63"/>
<Constant symbol="teta" name="scaling factor for the Wnt loop" value="1.12"/>
<Annotation>ODEs for the Wnt Loop</Annotation>
</System>
<Constant symbol="Wnt" value="C_12w*mFgf"/>
<Constant symbol="C_12w" name="Factor relating Wnt concentration and fgf mRNA concentration" value="0.32"/>
<CellDeath name="Leave the lattice at posterior boundary">
<Condition>cell.center.y < cD</Condition>
</CellDeath>
</CellType>
<CellType name="Wall" class="biological">
<FreezeMotion>
<Condition>1</Condition>
</FreezeMotion>
<ChangeCellType newCellType="Wall_Off" name="to Wall_Off">
<Condition>(cell.center.y >= wallDeath +5) + (cell.center.y < cD)*(wallDeath < 1.5*cD)</Condition>
</ChangeCellType>
<Constant symbol="mFgf" value="0"/>
<Constant symbol="F" name="lunatic fringe protein" value="0"/>
</CellType>
<CellType name="Wall_Off" class="biological">
<FreezeMotion>
<Condition>1</Condition>
</FreezeMotion>
<Property symbol="ticker" value="1"/>
<System time-step="1.0" solver="Euler [fixed, O(1)]">
<Rule symbol-ref="ticker">
<Expression>ticker-1</Expression>
</Rule>
</System>
<CellDeath>
<Condition>ticker <= 0 </Condition>
</CellDeath>
<Constant symbol="mFgf" value="0"/>
<Constant symbol="F" name="lunatic fringe protein" value="0"/>
</CellType>
<CellType name="preEphA4" class="biological">
<VolumeConstraint target="tV" strength="LamV"/>
<SurfaceConstraint target="tS" strength="LamS" mode="surface"/>
<Mapper time-step="1.0" name="sense FGF8 at cell center">
<Input value="if((space.x-1<=cell.center.x && space.x+1>=cell.center.x && space.y-1<=cell.center.y && space.y+1>=cell.center.y),FGF,0)"/>
<Output symbol-ref="FGF_center" mapping="maximum"/>
</Mapper>
<Property symbol="FGF_center" value="45"/>
<Property symbol="DL_m" name="delta protein in the membrane" value="0.0906"/>
<Property symbol="DL_mExt" name="Delta outside the cell" value="DL_m"/>
<Property symbol="F" name="lunatic fringe protein" value="0.0006"/>
<NeighborhoodReporter time-step="1.0" name="Delta/Notch from Neighboring cells">
<Input scaling="cell" value="DL_m"/>
<Output symbol-ref="DL_mExt" mapping="average"/>
</NeighborhoodReporter>
<Property symbol="mFgf" name="FGF8 mRNA" value="5"/>
<System time-step="1.0" name="FGF8" solver="Runge-Kutta [fixed, O(4)]">
<DiffEqn symbol-ref="mFgf" name="Decay, only for PSM cells">
<Expression>-k_fgf_cell*mFgf</Expression>
</DiffEqn>
</System>
<Property symbol="Wnt" value="C_12w*mFgf"/>
<Constant symbol="C_12w" name="Factor relating Wnt concentration and fgf mRNA concentration" value="0.32"/>
<System time-step="1.0" name="Wnt" solver="Euler [fixed, O(1)]">
<Rule symbol-ref="Wnt" name="Wnt decay">
<Expression>C_12w*mFgf</Expression>
</Rule>
</System>
<ChangeCellType newCellType="EphA4" name="to EphA4">
<Condition>ticker < 1</Condition>
<Triggers name="set_wall_death">
<Rule symbol-ref="wallDeath">
<Expression>cell.center.y</Expression>
</Rule>
</Triggers>
</ChangeCellType>
<Property symbol="ticker" name="determiantion ticker" value="24000"/>
<System time-step="1.0" name="determination ticker" solver="Euler [fixed, O(1)]">
<Rule symbol-ref="ticker">
<Expression>ticker-1</Expression>
</Rule>
</System>
</CellType>
<CellType name="preEphrinB2" class="biological">
<VolumeConstraint target="tV" strength="LamV"/>
<SurfaceConstraint target="tS" strength="LamS" mode="surface"/>
<Mapper time-step="1.0" name="sense FGF8 at cell center">
<Input value="if((space.x-1<=cell.center.x && space.x+1>=cell.center.x && space.y-1<=cell.center.y && space.y+1>=cell.center.y),FGF,0)"/>
<Output symbol-ref="FGF_center" mapping="maximum"/>
</Mapper>
<Property symbol="FGF_center" value="45"/>
<Property symbol="DL_m" name="delta protein in the membrane" value="0.0906"/>
<Property symbol="DL_mExt" name="Delta outside the cell" value="DL_m"/>
<Property symbol="F" name="lunatic fringe protein" value="0.0006"/>
<Property symbol="Sum_DL_mExt" name="Delta outside the cell" value="DL_m"/>
<Property symbol="PSM_Neighbors" name="Delta outside the cell" value="1"/>
<NeighborhoodReporter time-step="1.0" name="Delta/Notch from Neighboring cells">
<Input scaling="cell" value="DL_m"/>
<Output symbol-ref="Sum_DL_mExt" mapping="sum"/>
</NeighborhoodReporter>
<NeighborhoodReporter time-step="1.0" name="Neighboring PSM cells">
<Input scaling="cell" value="(cell.type == celltype.Source.id) + (cell.type == celltype.PSM.id) + (cell.type == celltype.preEphA4.id) + (cell.type == celltype.preCore.id) + (cell.type == celltype.preEphrinB2.id)"/>
<Output symbol-ref="PSM_Neighbors" mapping="sum"/>
</NeighborhoodReporter>
<System time-step="1.0" name="Delta/Notch signaling" solver="Euler [fixed, O(1)]">
<Rule symbol-ref="DL_mExt">
<Expression>if(PSM_Neighbors != 0, Sum_DL_mExt/PSM_Neighbors, 0)</Expression>
</Rule>
</System>
<Property symbol="mFgf" name="FGF8 mRNA" value="5"/>
<System time-step="1.0" name="FGF8" solver="Runge-Kutta [fixed, O(4)]">
<DiffEqn symbol-ref="mFgf" name="Decay, only for PSM cells">
<Expression>-k_fgf_cell*mFgf</Expression>
</DiffEqn>
</System>
<Property symbol="Wnt" value="C_12w*mFgf"/>
<Constant symbol="C_12w" name="Factor relating Wnt concentration and fgf mRNA concentration" value="0.32"/>
<System time-step="1.0" name="Wnt" solver="Euler [fixed, O(1)]">
<Rule symbol-ref="Wnt">
<Expression>C_12w*mFgf</Expression>
</Rule>
</System>
<ChangeCellType newCellType="EphrinB2" name="to EphrinB2">
<Condition>ticker < 1</Condition>
<Triggers name="set_wall_death">
<Rule symbol-ref="wallDeath">
<Expression>cell.center.y</Expression>
</Rule>
</Triggers>
</ChangeCellType>
<Property symbol="ticker" name="determiantion ticker" value="24000"/>
<System time-step="1.0" name="determination ticker" solver="Euler [fixed, O(1)]">
<Rule symbol-ref="ticker">
<Expression>ticker-1</Expression>
</Rule>
</System>
</CellType>
<CellType name="preCore" class="biological">
<VolumeConstraint target="tV" strength="LamV"/>
<SurfaceConstraint target="tS" strength="LamS" mode="surface"/>
<Mapper time-step="1.0" name="sense FGF8 at cell center">
<Input value="if((space.x-1<=cell.center.x && space.x+1>=cell.center.x && space.y-1<=cell.center.y && space.y+1>=cell.center.y),FGF,0)"/>
<Output symbol-ref="FGF_center" mapping="maximum"/>
</Mapper>
<Property symbol="FGF_center" value="45"/>
<Property symbol="DL_m" name="delta protein in the membrane" value="0.0906"/>
<Property symbol="DL_mExt" name="Delta outside the cell" value="DL_m"/>
<Property symbol="F" name="lunatic fringe protein" value="0.0006"/>
<Property symbol="Sum_DL_mExt" name="Delta outside the cell" value="DL_m"/>
<Property symbol="PSM_Neighbors" name="Delta outside the cell" value="1"/>
<NeighborhoodReporter time-step="1.0" name="Delta/Notch from Neighboring cells">
<Input scaling="cell" value="DL_m"/>
<Output symbol-ref="Sum_DL_mExt" mapping="sum"/>
</NeighborhoodReporter>
<NeighborhoodReporter time-step="1.0" name="Neighboring PSM cells">
<Input scaling="cell" value="(cell.type == celltype.Source.id) + (cell.type == celltype.PSM.id) + (cell.type == celltype.preEphA4.id) + (cell.type == celltype.preCore.id) + (cell.type == celltype.preEphrinB2.id)"/>
<Output symbol-ref="PSM_Neighbors" mapping="sum"/>
</NeighborhoodReporter>
<System time-step="1.0" name="Delta/Notch signaling" solver="Euler [fixed, O(1)]">
<Rule symbol-ref="DL_mExt">
<Expression>if(PSM_Neighbors != 0, Sum_DL_mExt/PSM_Neighbors, 0)</Expression>
</Rule>
</System>
<Property symbol="mFgf" name="FGF8 mRNA" value="5"/>
<System time-step="1.0" name="FGF8" solver="Runge-Kutta [fixed, O(4)]">
<DiffEqn symbol-ref="mFgf" name="Decay, only for PSM cells">
<Expression>-k_fgf_cell*mFgf</Expression>
</DiffEqn>
</System>
<Property symbol="Wnt" value="C_12w*mFgf"/>
<Constant symbol="C_12w" name="Factor relating Wnt concentration and fgf mRNA concentration" value="0.32"/>
<System time-step="1.0" name="Wnt" solver="Euler [fixed, O(1)]">
<Rule symbol-ref="Wnt" name="Wnt decay">
<Expression>C_12w*mFgf</Expression>
</Rule>
</System>
<ChangeCellType newCellType="Core" name="to Core">
<Condition>ticker < 1</Condition>
<Triggers name="set_wall_death">
<Rule symbol-ref="wallDeath">
<Expression>cell.center.y</Expression>
</Rule>
</Triggers>
</ChangeCellType>
<Property symbol="ticker" name="determiantion ticker" value="24000"/>
<System time-step="1.0" name="determination ticker" solver="Euler [fixed, O(1)]">
<Rule symbol-ref="ticker">
<Expression>ticker-1</Expression>
</Rule>
</System>
</CellType>
<CellType name="EphA4" class="biological">
<VolumeConstraint target="tV" strength="LamV"/>
<SurfaceConstraint target="tS" strength="LamS" mode="surface"/>
<Constant symbol="mFgf" value="0.0"/>
<Property symbol="F" name="lunatic fringe protein" value="0.0006"/>
</CellType>
<CellType name="EphrinB2" class="biological">
<VolumeConstraint target="tV" strength="LamV"/>
<SurfaceConstraint target="tS" strength="LamS" mode="surface"/>
<Constant symbol="mFgf" value="0.0"/>
<Property symbol="F" name="lunatic fringe protein" value="0.0006"/>
</CellType>
<CellType name="Core" class="biological">
<VolumeConstraint target="tV" strength="LamV"/>
<SurfaceConstraint target="tS" strength="LamS" mode="surface"/>
<Constant symbol="mFgf" value="0.0"/>
<Property symbol="F" name="lunatic fringe protein" value="0.0006"/>
</CellType>
<CellType name="PSM" class="biological">
<VolumeConstraint target="tV" strength="LamV"/>
<SurfaceConstraint target="tS" strength="LamS" mode="surface"/>
<Property symbol="FGF_center" value="45"/>
<Property symbol="N" name="Notch in the membrane" value="0.1144"/>
<Property symbol="N_a" name="activated Notch" value="0.7745"/>
<Property symbol="N_an" name="Activated Notch in the nucleus" value="0.00243"/>
<Property symbol="N_ap" name="phosphorylated notch" value="0.0013"/>
<Property symbol="MF" name="lunatic fringe mRNA" value="1.2464"/>
<Property symbol="F" name="lunatic fringe protein" value="2.3795"/>
<Property symbol="MDMF" name="delta modification factor mRNA" value="0.0011"/>
<Property symbol="DMF" name="delta modification factor protein" value="0.2090"/>
<Property symbol="DL_c" name="delta protein in cytoplasm" value="0.1255"/>
<Property symbol="DL_m" name="delta protein in the membrane" value="0.0906"/>
<Property symbol="DL_mExt" name="Delta outside the cell" value="DL_m"/>
<Property symbol="Sum_DL_mExt" name="Delta outside the cell" value="DL_m"/>
<Property symbol="PSM_Neighbors" name="Delta outside the cell" value="1"/>
<NeighborhoodReporter time-step="1.0" name="Delta/Notch from Neighboring cells">
<Input scaling="cell" value="DL_m"/>
<Output symbol-ref="Sum_DL_mExt" mapping="sum"/>
</NeighborhoodReporter>
<NeighborhoodReporter time-step="1.0" name="Neighboring PSM cells">
<Input scaling="cell" value="(cell.type == celltype.Source.id) + (cell.type == celltype.PSM.id) + (cell.type == celltype.preEphA4.id) + (cell.type == celltype.preCore.id) + (cell.type == celltype.preEphrinB2.id)"/>
<Output symbol-ref="PSM_Neighbors" mapping="sum"/>
</NeighborhoodReporter>
<System time-step="1.0" name="Delta/Notch signaling" solver="Euler [fixed, O(1)]">
<Rule symbol-ref="DL_mExt">
<Expression>if(PSM_Neighbors != 0, Sum_DL_mExt/PSM_Neighbors, 0)</Expression>
</Rule>
</System>
<System time-step="1.0" name="Notch loop" solver="Runge-Kutta [fixed, O(4)]">
<DiffEqn symbol-ref="N">
<Expression>eps*(mu_sN-mu_dN*N/(K_dN+N)-(k_c*N*(K_IF^2/(K_IF^2 + F^2))*((DL_mExt/(K_DL +DL_mExt))/(K_aDL + (DL_mExt/(K_DL +DL_mExt))))))</Expression>
</DiffEqn>
<DiffEqn symbol-ref="N_a">
<Expression>eps*((k_c*N*(K_IF^2/(K_IF^2 + F^2))*((DL_mExt/(K_DL +DL_mExt))/(K_aDL + (DL_mExt/(K_DL +DL_mExt)))))-mu_dNa*N_a/(K_dNa +N_a)-(k_t1*N_a-k_t2*N_an))</Expression>
</DiffEqn>
<DiffEqn symbol-ref="N_an">
<Expression>eps*((k_t1*N_a-k_t2*N_an)-mu_dNan*N_an/(K_dNan + N_an))</Expression>
</DiffEqn>
<DiffEqn symbol-ref="MF">
<Expression>eps*(mu_sF*(N_an-(mu_Nap*N_an*K/(K_pN+K)))^2/(KA^2 +(N_an-(mu_Nap*N_an*K/(K_pN+K)))^2)-mu_mF*MF/(K_dmF+MF))</Expression>
</DiffEqn>
<DiffEqn symbol-ref="F">
<Expression>eps*(k_sF*MF -mu_dF*F/(K_dF+F))</Expression>
</DiffEqn>
<DiffEqn symbol-ref="MDMF">
<Expression>eps*(mu_s0MDMF*(N_an-(mu_Nap*N_an*K/(K_pN+K)))^2/(K_aNDMF^2+(N_an-(mu_Nap*N_an*K/(K_pN+K)))^2) - mu_dmDMF*MDMF/(K_dmDMF +MDMF))</Expression>
</DiffEqn>
<DiffEqn symbol-ref="DMF">
<Expression>eps*(k_sDMF*MDMF - mu_dDMF*DMF/(K_dDMF + DMF))</Expression>
</DiffEqn>
<DiffEqn symbol-ref="DL_c">
<Expression>eps*(k_sDL*MDL*(N_an -(mu_Nap*N_an*K/(K_pN+K)))/(K_Nan +(N_an-(mu_Nap*N_an*K/(K_pN+K)))) -k_tDL*DL_c - mu_dDLc*DL_c/(K_dDLc + DL_c))</Expression>
</DiffEqn>
<DiffEqn symbol-ref="DL_m">
<Expression>eps*(k_tDL*DL_c - mu_dDLm*DL_m/(K_dDLm + DL_m))</Expression>
</DiffEqn>
<Constant symbol="mu_sN" name="maximum rate of notch synthesis" value="0.23*mpm"/>
<Constant symbol="mu_dN" name="maximum rate of notch degradation" value="2.82*mpm"/>
<Constant symbol="K_dN" name="michaelis constant for notch degradation" value="1.4"/>
<Constant symbol="k_c" name="rate for notch cleavage" value="3.45*mpm"/>
<Constant symbol="mu_dNa" name="maximum rate of cytoplasmic NICD degradation" value="0.01*mpm"/>
<Constant symbol="K_dNa" name="michaelis constant for cytoplasmic NICD degradation" value="0.001"/>
<Constant symbol="mu_dNan" name="maximum rate of nuclear NICD degradation" value="0.1*mpm"/>
<Constant symbol="K_dNan" value="0.001"/>
<Constant symbol="K_IF" name="threshold constant for inhibition of notch cleavage by Lfng" value="0.45"/>
<Constant symbol="K_aDL" name="delta signaling threshold constant for notch cleavage" value="0.035"/>
<Constant symbol="k_t1" name="rate constant for NICD entry into nucleus" value="0.1*mpm"/>
<Constant symbol="k_t2" value="0.1*mpm"/>
<Constant symbol="mu_Nap" name="maximum fraction of NICD phosphorylated" value="1.0"/>
<Constant symbol="K_pN" name="Threshold constant for Gsk3Beta-mediated NICD phosphorylation" value="2.5"/>
<Constant symbol="mu_sF" name="maximum rate of Lfng transcription" value="3.24*mpm"/>
<Constant symbol="K_A" name="threshold constant for activation of Lfng gene by NICD" value="0.05"/>
<Constant symbol="mu_mF" name="maximum rate of Lfng mRNA degradation" value="1.92*mpm"/>
<Constant symbol="K_dmF" name="michaelis constant for Lfng mRNA degradation" value="0.768"/>
<Constant symbol="k_sF" name="rate constant for Lfng protein synthesis" value="0.3*mpm"/>
<Constant symbol="mu_dF" name="maximum rate of Lfng protein degradation" value="0.39*mpm"/>
<Constant symbol="K_dF" name="michaelis constant for Lfng protein degradation" value="0.37"/>
<Constant symbol="mu_s0MDMF" name="maximum rate of DMF transcription" value="0.497*mpm"/>
<Constant symbol="K_aNDMF" name="michaelis constant for notch activation of DMF" value="0.05"/>
<Constant symbol="mu_dmDMF" name="maximum rate of DMF mRNA degradation" value="0.314*mpm"/>
<Constant symbol="K_dmDMF" name="michaelis constant for DMD mRNA degradation" value="0.4"/>
<Constant symbol="k_sDMF" name="rate constant for translation of DMF" value="0.1047*mpm"/>
<Constant symbol="mu_dDMF" name="maximum rate of DMF protein degradation" value="0.209*mpm"/>
<Constant symbol="K_dDMF" name="michaelis constant for DMF protein degradation" value="0.5"/>
<Constant symbol="k_sDL" name="rate constant for delta protein synthesis" value="0.75*mpm"/>
<Constant symbol="MDL" name="concentration of delta mRNA" value="0.5"/>
<Constant symbol="K_Nan" name="michaelis constant for NICD stimulation of delta translation" value="0.04"/>
<Constant symbol="k_tDL" name="rate constant for delta protein transport to membrane" value="0.5*mpm"/>
<Constant symbol="mu_dDLc" name="maximum rate of delta proteind egradation in cytoplasm" value="0.5*mpm"/>
<Constant symbol="K_dDLc" name="michaelis constant for delta protein degradation in cytoplasm" value="0.5"/>
<Constant symbol="mu_dDLm" name="maximum rate of delta protein degradation on membrane" value="0.5*mpm"/>
<Constant symbol="K_dDLm" name="michaelis constant for delta protein degradation on membrane" value="0.5"/>
<Constant symbol="K_DL" name="Threshold for delta signaling" value="0.08"/>
<Constant symbol="eps" name="scaling factor for notch loop" value="0.43"/>
<Constant symbol="KA" value="0.05"/>
<Annotation>ODEs for the Notch Loop</Annotation>
</System>
<Property symbol="Ras_a" name="Activated Ras protein" value="1.9888"/>
<Property symbol="ERK_a" name="Activated ERK protein" value="0.01930"/>
<Property symbol="X_a" name="Activated transcription factor X" value="1.9777"/>
<Property symbol="MDusp" name="Dusp mRNA" value="2.1651"/>
<Property symbol="Dusp" name="Dusp protein" value="8.5078"/>
<Property symbol="mFgf" name="FGF8 mRNA" value="if((size.y-4*cD-cell.center.y>0)*(size.y-4*cD-cell.center.y<=cD), 4.4, if((size.y-4*cD-cell.center.y>cD)*(size.y-4*cD-cell.center.y<=(cD)*2), 4.6, if((size.y-4*cD-cell.center.y>(cD)*2), 4.8, 4.8)))"/>
<System time-step="1.0" name="FGF loop" solver="Runge-Kutta [fixed, O(4)]">
<DiffEqn symbol-ref="Ras_a">
<Expression>eta*(V_MaRas*((FGF_center^2/(K_aFgf^2 + FGF_center^2))*((Ras_t-Ras_a)/(K_aRas + (Ras_t-Ras_a)))-V_MdRas*(Ras_a/(K_dRas+Ras_a))))</Expression>
</DiffEqn>
<DiffEqn symbol-ref="ERK_a">
<Expression>eta*(V_MaErk*(Ras_a/Ras_t)*(ERK_t-ERK_a)/(K_aErk + (ERK_t-ERK_a))-K_cDusp*Dusp*ERK_a/(K_dErk+ERK_a))</Expression>
</DiffEqn>
<DiffEqn symbol-ref="X_a">
<Expression>eta*(V_MaX*ERK_a/ERK_t*(X_t-X_a)/(K_aX +(X_t-X_a))-V_MdX*X_a/(K_dX + X_a))</Expression>
</DiffEqn>
<DiffEqn symbol-ref="MDusp">
<Expression>eta*(V_MsMDusp*(X_a^2/(K_aMDusp^2 + X_a^2))*(mu_DuspDMF*K_IMDusp/(K_IMDusp +DMF) +mu_DuspX) - V_MdMDusp*MDusp/(K_dMDusp +MDusp))</Expression>
</DiffEqn>
<DiffEqn symbol-ref="Dusp">
<Expression>eta*(K_sDusp*MDusp-V_dDusp*Dusp/(K_dDusp + Dusp))</Expression>
</DiffEqn>
<DiffEqn symbol-ref="mFgf" name="Decay, only for PSM cells">
<Expression>-k_fgf_cell*mFgf</Expression>
</DiffEqn>
<Constant symbol="Ras_t" name="Total concentration of Ras protein" value="2.0"/>
<Constant symbol="V_MaRas" name="Maximum rate of Ras activation" value="4.968*mpm"/>
<Constant symbol="K_aFgf" name="FGF8 threshold constant for activation of Ras" value="0.5"/>
<Constant symbol="K_aRas" name="Inactie Ras threshold constant for activation of Ras" value="0.103"/>
<Constant symbol="V_MdRas" name="Maximum rate of Ras activation" value="0.41*mpm"/>
<Constant symbol="K_dRas" name="Michaelis constant of Ras activation" value="0.1"/>
<Constant symbol="ERK_t" name="Total concentration of ERK protein kinase" value="2.0"/>
<Constant symbol="V_MaErk" name="Maximum rate of Ras-mediated ERK activation" value="3.3*mpm"/>
<Constant symbol="K_aErk" name="Inactive ERK threshold constant for ERK activation" value="0.05"/>
<Constant symbol="K_cDusp" name="Rate constant for inactivation of ERK" value="1.35*mpm"/>
<Constant symbol="K_dErk" name="Michaelis constant for inactivation of ERK" value="0.05"/>
<Constant symbol="X_t" name="Total concentration of factor X" value="2.0"/>
<Constant symbol="V_MaX" name="Maximum rate of ERK-activated X activation" value="1.6*mpm"/>
<Constant symbol="K_aX" name="Threshold constant for ERK-mediated X activation" value="0.05"/>
<Constant symbol="V_MdX" name="Maximum rate of X inactivation" value="0.5*mpm"/>
<Constant symbol="K_dX" name="Michaelis constant for X inactivation" value="0.05"/>
<Constant symbol="V_MsMDusp" name="maximum rate of Dusp6 transcription" value="0.9*mpm"/>
<Constant symbol="K_aMDusp" name="Threshold constant for X-activated Dusp6-transcription" value="0.5"/>
<Constant symbol="V_MdMDusp" name="maximum rate of Dusp6 mRNA degradation" value="0.5*mpm"/>
<Constant symbol="K_dMDusp" name="Michaelis constant for Dusp6 mRNA degradation" value="0.5"/>
<Constant symbol="K_sDusp" name="rate constant for Dusp6 translation" value="0.5*mpm"/>
<Constant symbol="V_dDusp" name="Maximum rate of Dusp6protein degradation" value="2.0*mpm"/>
<Constant symbol="K_dDusp" name="Michaelis constant for Dusp6 protein degradation" value="0.5"/>
<Constant symbol="mu_DuspX" name="Fraction of Dusp6 transcription solely regulated by X" value="0.2"/>
<Constant symbol="mu_DuspDMF" name="Fraction of Dusp6 transcription under regulation by DMF" value="0.8"/>
<Constant symbol="K_IMDusp" name="Threshold constant for DMF-mediated inhibition of Dusp6" value="0.3"/>
<Constant symbol="eta" name="Scaling factor for FGF8 loop" value="0.328"/>
<Annotation>ODEs for the FGF Loop</Annotation>
</System>
<Property symbol="K" name="Free Gsk3 protein" value="1.4428"/>
<Property symbol="B" name="Beta-Katenin" value="0.4705"/>
<Property symbol="B_p" name="Phosphorylated beta catenin" value="0.0090"/>
<Property symbol="B_n" value="0.2146"/>
<Property symbol="MAx" name="Axin2 mRNA" value="0.1523"/>
<Property symbol="A" name="Axin2 protein" value="0.0461"/>
<System time-step="1.0" name="Wnt loop" solver="Runge-Kutta [fixed, O(4)]">
<DiffEqn symbol-ref="K">
<Expression>teta*(d_1*(K_t-K)-a_1*A*K)</Expression>
</DiffEqn>
<DiffEqn symbol-ref="B">
<Expression>teta*(mu_sB-(V_MK*K_ID/(K_ID+Wnt)*B/(K_1+B))*(K_t-K)/K_t+(V_MP*B_p/(K_2+B_p))+(k_t4*B_n-k_t3*B)-k_d1*B)</Expression>
</DiffEqn>
<DiffEqn symbol-ref="B_p">
<Expression>teta*((V_MK*K_ID/(K_ID+Wnt)*B/(K_1+B))*(K_t-K)/K_t-(V_MP*B_p/(K_2+B_p))-k_d2*B_p)</Expression>
</DiffEqn>
<DiffEqn symbol-ref="B_n">
<Expression>-teta*(k_t4*B_n-k_t3*B)</Expression>
</DiffEqn>
<DiffEqn symbol-ref="MAx">
<Expression>teta*(mu_0+mu_MB*B_n^2/(K_aB^2+B_n^2)+mu_MXa*X_a^2/(K_aXa^2+X_a^2)-mu_md*MAx/(K_md+MAx))</Expression>
</DiffEqn>
<DiffEqn symbol-ref="A">
<Expression>teta*(k_sAx*MAx-mu_dAx*A/(K_dAx+A)+(d_1*(K_t-K)-a_1*A*K))</Expression>
</DiffEqn>
<Constant symbol="a_1" name="Rate constant for Gsk3-Axin-Binding" value="1.8*mpm"/>
<Constant symbol="d_1" name="Rate constant for Gsk3-Axin dissociation" value="0.1*mpm"/>
<Constant symbol="mu_sB" name="Maximum rate of beta-catenin synthesis" value="0.087*mpm"/>
<Constant symbol="k_t3" name="Rate constant for beta-catenin entry into nucleus" value="0.7*mpm"/>
<Constant symbol="k_t4" name="Rate constant for beta-catenin exit from nucleus" value="1.5*mpm"/>
<Constant symbol="V_MK" name="Maximum rate of Gsk3Beta-medaited beta-catein phosphorylation" value="4.5*mpm"/>
<Constant symbol="K_t" name="Total Gsk3Beta concentration" value="3.0"/>
<Constant symbol="K_ID" name="Threshold constant for Dsh inhibition of beta-catenin phosphorylation" value="0.5"/>
<Constant symbol="K_1" name="Michaelis constant for beta-catenin phosphorylation" value="0.28"/>
<Constant symbol="V_MP" name="Maximum rate of beta-catenin dephosphorylation" value="1.0*mpm"/>
<Constant symbol="K_2" name="Michaelis constant for b-catenin dephosphorylation" value="0.03 "/>
<Constant symbol="k_d1" name="Rate constant for unphosphorylated b-catenin degradation" value="0.0*mpm"/>
<Constant symbol="k_d2" name="rate constant for phosphorylated beta-catenin degradation" value="7.062*mpm"/>
<Constant symbol="mu_0" name="Basal rate of Axin2 transcription" value="0.06*mpm"/>
<Constant symbol="mu_MB" name="Maximum rate of beta-catenin activated Axin2 transcription" value="1.64*mpm"/>
<Constant symbol="K_aB" name="Threshold constant for beta-catenon activation of Axin2" value="0.7"/>
<Constant symbol="mu_md" name="Maximum rate of Axin2 mRNA degradation" value="0.8*mpm"/>
<Constant symbol="K_md" name="Michaelis constant for Axin2 mRNA degradation" value="0.48"/>
<Constant symbol="mu_MXa" name="Maximum rate of Xa activated Axin2 transcription" value="0.5*mpm"/>
<Constant symbol="K_aXa" name="Threshold constant for Xa activation of Axin2" value="0.05"/>
<Constant symbol="k_sAx" name="rate constant for axin2 translation" value="0.02*mpm"/>
<Constant symbol="mu_dAx" name="Maximum rate of Axin2 protein degradation" value="0.6*mpm"/>
<Constant symbol="K_dAx" name="Michaelis constant for Axin2 protein degradation" value="0.63"/>
<Constant symbol="teta" name="scaling factor for the Wnt loop" value="1.12"/>
<Annotation>ODEs for the Wnt Loop</Annotation>
</System>
<Property symbol="Wnt" value="C_12w*mFgf"/>
<Constant symbol="C_12w" name="Factor relating Wnt concentration and fgf mRNA concentration" value="0.32"/>
<System time-step="1.0" name="Wnt decay" solver="Euler [fixed, O(1)]">
<Rule symbol-ref="Wnt">
<Expression>C_12w*mFgf</Expression>
<Annotation>Equation for Wnt degredation</Annotation>
</Rule>
</System>
<Mapper time-step="1.0" name="Sense FGF8 at cell center">
<Input value="if((space.x-1<=cell.center.x && space.x+1>=cell.center.x && space.y-1<=cell.center.y && space.y+1>=cell.center.y),FGF,0)"/>
<Output symbol-ref="FGF_center" mapping="maximum"/>
</Mapper>
<ChangeCellType newCellType="preCore" name="to preCore">
<Condition>(FGF_center < fdTh)*(FGF_center > 0.000001)*(B>bdTh)</Condition>
<Annotation>Change cell type as soon as a threshold of fgf concentration is reached. If the beta catenenin concentration is above another threshold, the PSM cell differentiates to a pre_core cell.</Annotation>
</ChangeCellType>
<ChangeCellType newCellType="preEphrinB2" name="to preEphrinB2">
<Condition>(FGF_center < fdTh)*(FGF_center > 0.000001)*(B<=bdTh)*(F<=A*aPre)</Condition>
<Annotation>Change cell type as soon as a threshold of fgf concentration is reached. If the beta catenenin concentration is below another threshold, and the Axin2 concentration times a factor is bigger or equal to the Lfng concentrentration the PSM cell differentiates to a pre_EphrinBB2 cell.</Annotation>
</ChangeCellType>
<ChangeCellType newCellType="preEphA4" name="to preEphA4">
<Condition>(FGF_center < fdTh)*(FGF_center > 0.000001)*(B<=bdTh)*(F>A*aPre)</Condition>
<Annotation>Change cell type as soon as a threshold of fgf concentration is reached. If the beta catenenin concentration is below another threshold, and the Axin2 concentration times a factor is smaller than the Lfng concentrentration the PSM cell differentiates to a pre_EphA4 cell.</Annotation>
</ChangeCellType>
</CellType>
<CellType name="Medium" class="medium">
<Constant symbol="mFgf" value="0"/>
<Constant symbol="F" name="lunatic fringe protein" value="0"/>
</CellType>
</CellTypes>
<CPM>
<Interaction>
<Contact type2="Medium" type1="Medium" value="0.0"/>
<Contact type2="PSM" type1="Medium" value="15.0"/>
<Contact type2="Source" type1="Medium" value="0.0"/>
<Contact type2="preEphA4" type1="Medium" value="15.0"/>
<Contact type2="preEphrinB2" type1="Medium" value="15.0"/>
<Contact type2="preCore" type1="Medium" value="15.0"/>
<Contact type2="EphA4" type1="Medium" value="5.0"/>
<Contact type2="EphrinB2" type1="Medium" value="5.0"/>
<Contact type2="Core" type1="Medium" value="15.0"/>
<Contact type2="Wall" type1="Medium" value="0.0"/>
<Contact type2="Wall_Off" type1="Medium" value="100"/>
<Contact type2="PSM" type1="PSM" value="-20.0"/>
<Contact type2="Source" type1="PSM" value="-20.0"/>
<Contact type2="preEphA4" type1="PSM" value="-20.0"/>
<Contact type2="preEphrinB2" type1="PSM" value="-20.0"/>
<Contact type2="preCore" type1="PSM" value="-20.0"/>
<Contact type2="EphA4" type1="PSM" value="-20.0"/>
<Contact type2="EphrinB2" type1="PSM" value="-20.0"/>
<Contact type2="Core" type1="PSM" value="-20.0"/>
<Contact type2="Wall" type1="PSM" value="30.0"/>
<Contact type2="Wall_Off" type1="PSM" value="100"/>
<Contact type2="preEphA4" type1="Source" value="-20.0"/>
<Contact type2="preEphrinB2" type1="Source" value="-20.0"/>
<Contact type2="preCore" type1="Source" value="-20.0"/>
<Contact type2="EphA4" type1="Source" value="-20.0"/>
<Contact type2="EphrinB2" type1="Source" value="-20.0"/>
<Contact type2="Core" type1="Source" value="-20.0"/>
<Contact type2="Wall" type1="Source" value="30.0"/>
<Contact type2="Wall_Off" type1="Source" value="100"/>
<Contact type2="Source" type1="Source" value="-20.0"/>
<Contact type2="preEphA4" type1="preEphA4" value="-25.0"/>
<Contact type2="preEphrinB2" type1="preEphA4" value="-20.0"/>
<Contact type2="preCore" type1="preEphA4" value="-20.0"/>
<Contact type2="EphA4" type1="preEphA4" value="-25.0"/>
<Contact type2="EphrinB2" type1="preEphA4" value="-20.0"/>
<Contact type2="Core" type1="preEphA4" value="-20.0"/>
<Contact type2="Wall" type1="preEphA4" value="30.0"/>
<Contact type2="Wall_Off" type1="preEphA4" value="100"/>
<Contact type2="preEphrinB2" type1="preEphrinB2" value="-25.0"/>
<Contact type2="preCore" type1="preEphrinB2" value="-20.0"/>
<Contact type2="EphA4" type1="preEphrinB2" value="-20.0"/>
<Contact type2="EphrinB2" type1="preEphrinB2" value="-25.0"/>
<Contact type2="Core" type1="preEphrinB2" value="-20.0"/>
<Contact type2="Wall" type1="preEphrinB2" value="30.0"/>
<Contact type2="Wall_Off" type1="preEphrinB2" value="100"/>
<Contact type2="preCore" type1="preCore" value="-35.0"/>
<Contact type2="EphA4" type1="preCore" value="-20.0"/>
<Contact type2="EphrinB2" type1="preCore" value="-20.0"/>
<Contact type2="Core" type1="preCore" value="-20.0"/>
<Contact type2="Wall" type1="preCore" value="30.0"/>
<Contact type2="Wall_Off" type1="preCore" value="100"/>
<Contact type2="EphA4" type1="EphA4" value="-25.0"/>
<Contact type2="EphrinB2" type1="EphA4" value="80.0"/>
<Contact type2="Core" type1="EphA4" value="-25.0"/>
<Contact type2="Wall" type1="EphA4" value="30.0"/>
<Contact type2="Wall_Off" type1="EphA4" value="100"/>
<Contact type2="EphrinB2" type1="EphrinB2" value="-25.0"/>
<Contact type2="Core" type1="EphrinB2" value="-25.0"/>
<Contact type2="Wall" type1="EphrinB2" value="30.0"/>
<Contact type2="Wall_Off" type1="EphrinB2" value="100"/>
<Contact type2="Core" type1="Core" value="-40.0"/>
<Contact type2="Wall" type1="Core" value="30.0"/>
<Contact type2="Wall_Off" type1="Core" value="100"/>
<Contact type2="Wall" type1="Wall" value="0.0"/>
<Contact type2="Wall_Off" type1="Wall" value="100"/>
<Contact type2="Wall_Off" type1="Wall_Off" value="100"/>
</Interaction>
<ShapeSurface scaling="classic">
<Neighborhood>
<Order>2</Order>
</Neighborhood>
</ShapeSurface>
<MonteCarloSampler stepper="edgelist">
<MCSDuration value="1"/>
<MetropolisKinetics temperature="120"/>
<Neighborhood>
<Order>4</Order>
</Neighborhood>
</MonteCarloSampler>
</CPM>
<CellPopulations>
<Population type="Wall" name="Two columns of Wall cells" size="0">
<InitCellObjects mode="distance">
<Arrangement repetitions="2, size.y/wall, 0" displacements="cD*11, wall, 0">
<Box origin="margin, 0.0, 0.0" size="wall,wall, 0.0"/>
</Arrangement>
</InitCellObjects>
</Population>
<Population type="PSM" name="PSM tissue" size="0">
<InitCellObjects mode="distance">
<Arrangement repetitions="10, 3, 0" displacements="cD, cD, 0">
<Box origin="margin + wall, size.y-18-4*cD, 0.0" size="cD, cD, 0.0"/>
</Arrangement>
</InitCellObjects>
</Population>
<Population type="Source" name="Source cell layer" size="0">
<InitCellObjects mode="distance">
<Arrangement repetitions="10, 1, 0" displacements="cD, 0, 0">
<Box origin="margin + wall, size.y-24-4*cD, 0.0" size="cD, cD, 0.0"/>
</Arrangement>
</InitCellObjects>
</Population>
</CellPopulations>
<Global>
<Constant symbol="cD" name="Cell diameter" value="7">
<Annotation>Default cell diameter</Annotation>
</Constant>
<Constant symbol="tS" name="Target surface" value="28">
<Annotation>Default target surface</Annotation>
</Constant>
<Constant symbol="LamS" name="Surface strength" value="15">
<Annotation>Default weight for CPM surface constraint, termed lambda_s in Hester et al. 2011</Annotation>
</Constant>
<Constant symbol="LamV" name="Volume strength" value="15">
<Annotation>Default weight for CPM volume constraint, termed lambda_v in Hester et al. 2011</Annotation>
</Constant>
<Constant symbol="tV" name="Target volume" value="cD*cD"/>
<Constant symbol="margin" name="Margin" value="35">
<Annotation>Number of empty lattice nodes between wall cells and lattice boundary</Annotation>
</Constant>
<Constant symbol="wall" name="Wall cell diameter" value="7">
<Annotation>Width of the two columns made by Wall cells</Annotation>
</Constant>
<Constant symbol="slopeFGF" name="Initial slope of FGF8 gradient" value="(45-39.6)/(cD*4)">
<Annotation>Initial slope of FGF8 gradient (used for initializing FGF field) computed as the value of FGF (from Hester et al. 2011, page 13 bottom right) at the posterior end (45nM) minus the value of FGF at the fourth cell layer (39.6 nM) divided by the thickness of 4 cell layers.</Annotation>
</Constant>
<Field symbol="FGF" value="if((size.y-4*cD-space.y <= cD*4 +3)*(size.y-4*cD-space.y>0)*(space.x >= margin + wall)*(space.x <= margin+ wall + 10*cD), 39.6 + slopeFGF*(size.y-4*cD-space.y), 0)">
<Diffusion rate="0.6*0.7*0.7*mpm"/>
<Annotation>Initialization of FGF8 gradient and diffusion term (units were rescaled from µm^2/min to pixel^2/MCS) as defined in Hester et al., 2011</Annotation>
</Field>
<System time-step="1" name="FGF gradient formation" solver="Heun [fixed, O(2)]">
<DiffEqn symbol-ref="FGF">
<Expression>-k_FGF_global*FGF + s_FGF*mFgf</Expression>
</DiffEqn>
<Constant symbol="k_FGF_global" name="FGF decay rate" value="0.2*mpm"/>
<Constant symbol="s_FGF" name="FGF secretion rate (gets multiplied by FGF mRNA from Source and PSM cells, hence becomes 0 elsewhere)" value="1.83*mpm"/>
</System>
<Constant symbol="mpm" name="Time unit of 0.015 min per 1 Monte Carlo step (MCS)" value="0.015">
<Annotation>Rates (and time scale of diffusion coefficient) were given in 1/min and need to be rescaled into 1/MCS using "1 MCS=mpm min -> 1 min=1/mpm MCS and 1/min = mpm 1/MCS" hence rates have been multiplied by mpm.</Annotation>
</Constant>
<Constant symbol="k_fgf_cell" name="Decay rate of FGF8 in PSM cells" value="0.005*mpm"/>
<Constant symbol="DL_m" name="Background Delta expression of passive cells (Wall)" value="0.0"/>
<Constant symbol="fdTh" name="FGF8 determination threshold" value="13.9"/>
<Constant symbol="bdTh" name="Beta catenin determination threshold" value="0.406"/>
<Constant symbol="aPre" name="Prefactor for Axin2 for determination threshold" value="21.28"/>
<Variable symbol="wallDeath" value="size.y + 10">
<Annotation>System-wide variable that specifies where somites have already formed, i.e. where the wall can disappear. Initialized with value outside of the lattice to maintain wall initially.</Annotation>
</Variable>
<Constant symbol="gR" name="Growth rate of target volume in source cells" value="0.12">
<Annotation> As specified in the code in Hester's Dissertation ("Grow=0.120" on page 129)</Annotation>
</Constant>
</Global>
</MorpheusModel>

Downloads
Files associated with this model: